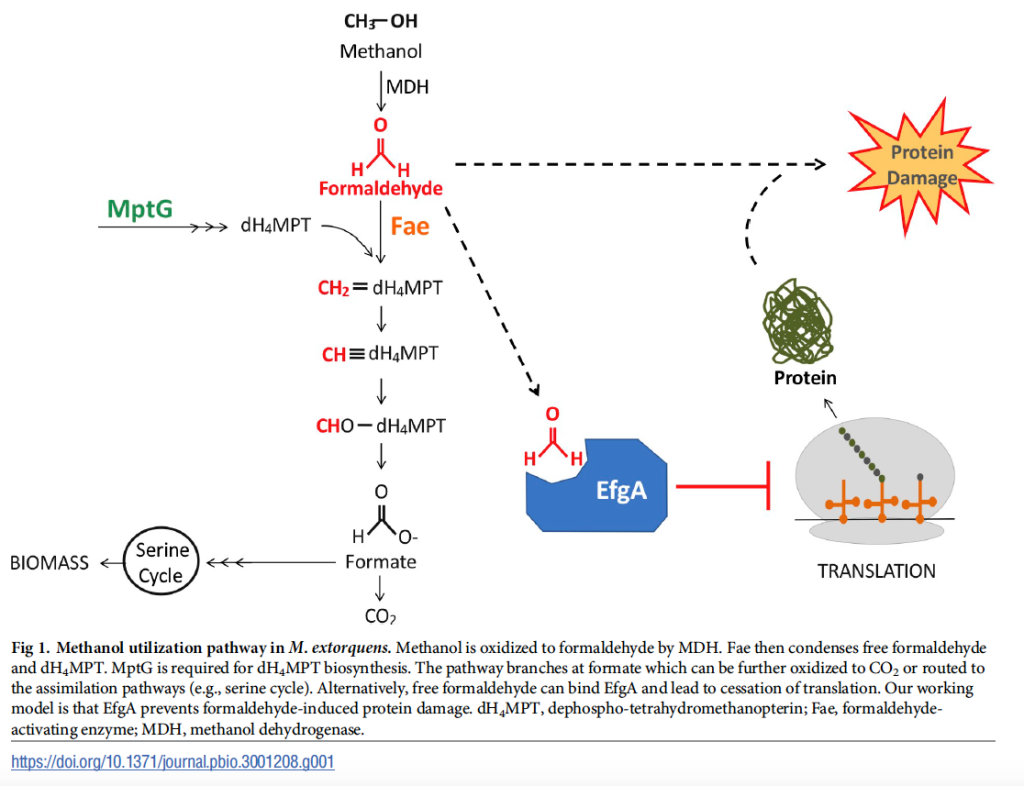
After a long period of work and many contributions from collaborators and many people within the lab our discovery of the formaldehyde sensor EfgA came out in a paper in PLOS Biology. This work, led by Jannell Bazurto (now faculty at University of Minnesota) and initiated by Dipti Nayak (now faculty at UC Berkeley) describes the surprising finding that methylotrophs possess a sensor for formaldehyde that appears to directly lead to growth cessation if levels of the potent toxin rise too high.
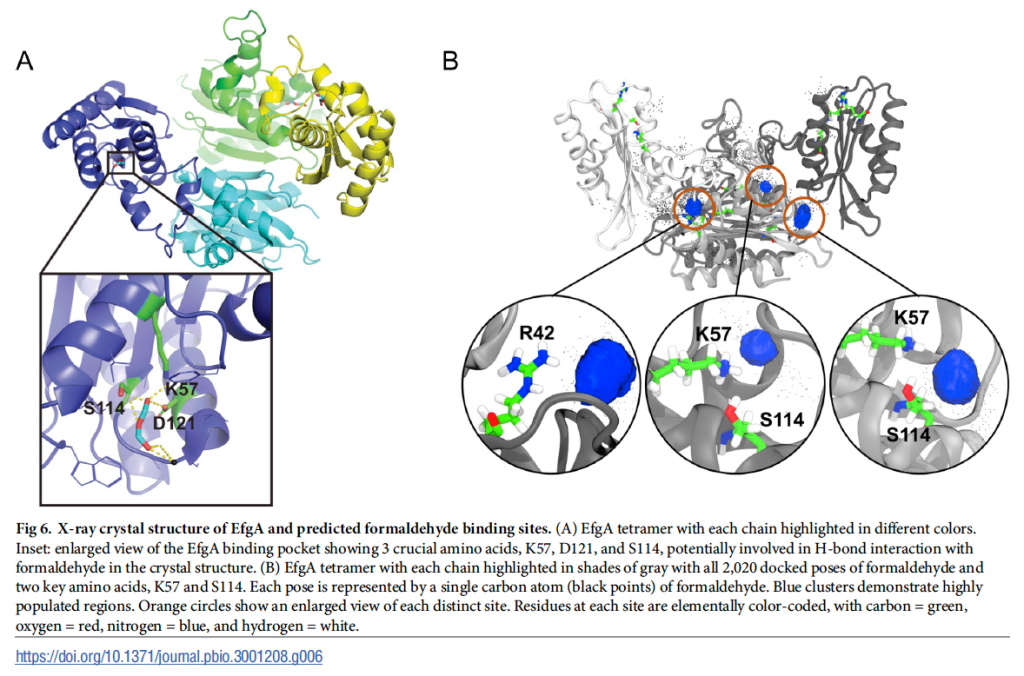
Collaboration with Yousif Shamoo at Rice University led to solving the crystal structure of EfgA and the formaldehyde binding pocket, a result supported by both computational work by Jagdish Patel and Marty Ytreberg here at University of Idaho, and biochemical experiments by a former postdoc, Tomislav Ticak, who is now a Scientist at Proctor & Gamble). Rather unexpectedly, introduction of EfgA from Methylorubrum extorquens into Escherichia coli resulted in an enhanced ability to grow on glucose in the presence of formaldehyde. This result, as well finding alternative loci for beneficial mutations that enabled growth on formaldehyde has led to the working model that EfgA binds formaldehyde and directly interacts with ribosomes to slow or shut down translation in response to elevated internal formaldehyde.
Two other recent papers by Jannell build upon this result. First, a paper in Journal of Bacteriology demonstrates that EfgA (as well as the MarR-like transcriptional regulator we name TtmR) is critical for the transition from multi-C substrates to C1 substrates like methanol. Second, a paper co-first authored by former PhD student Siavash Riazi and in collaboration with Jeff Barrick at UT Austin in Microorganisms shows that the global transcriptional response of cells to formaldehyde bears many similarities to exposure to a translational inhibitor (kanamycin).
All of this goes to show that methylotrophs possess more than just the enzymes to convert C1 substrates into energy and central metabolites, but that they also possess an intricate system of regulators that appear to work at both transcriptional and translational levels to deal with the toxicity encountered dealing with formaldehyde as a central metabolite.
Finally, there are a couple nice press releases put out by Rice and PLOS (on EurekaAlert, AAAS) on this work. Thanks so much for these.
