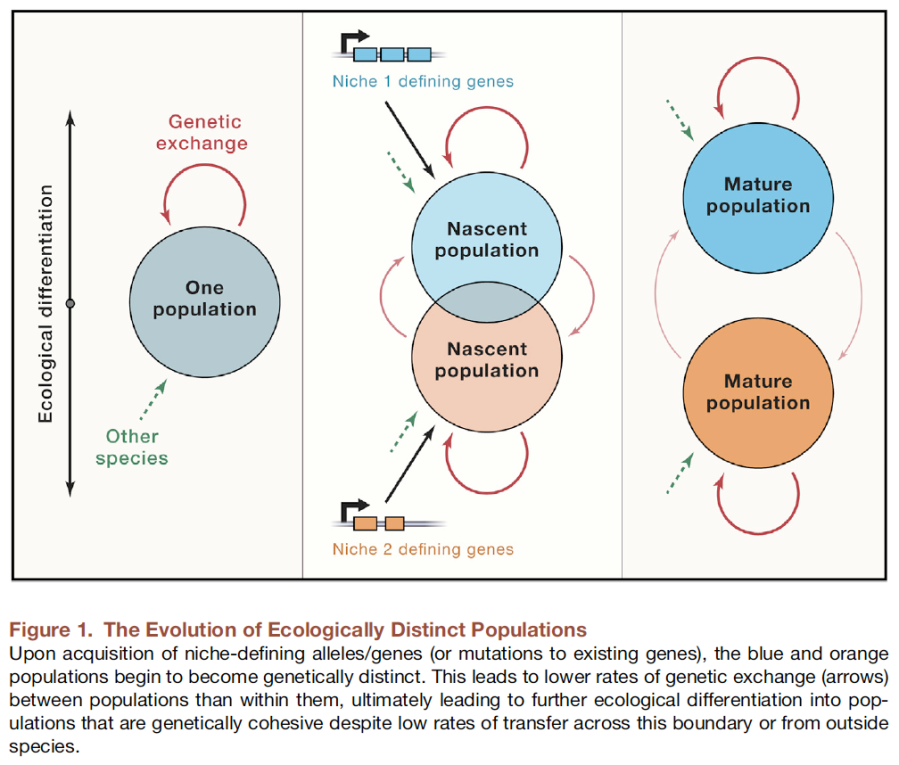
In the morass of metagenomic sequence data on microbes, it is unclear what a unit should even be in a community. Ideally, one would want to uncover the core populations present: the set of strains that actually are exchanging genetic information and are reasonably cohesive, even in the presence of a degree of input from outside sources. The Polz lab has put forward a fantastic new method in Arevalo et al. “A reverse ecology approach based on a biological definition of microbial populations” (Cell, 2019. 178:820-834). for doing so that is based upon the simple concept that the length distribution of perfectly matching sequences can serve as a chronometer of separation. In the absence of exchange, two lineages emerging from the same genome sequence will ultimately accumulate random differences that will leave behind an exponential distribution of perfect sequence matches. If they continue to exchange, however, this will lead to an excess of longer sequence matches than expected. Indeed, this pattern is seen amongst strains previously identified as being of the same population. Furthermore, there is the substantial advantage that it identifies precisely which genes are being exchanged, thereby providing clues as to the success of one population versus each other, hence the concept of a “reverse” approach to ecology.
In our Preview – “Align to define: ecologically meaningful populations from genomes” (Cell, 2019. 178:767-768) – Sergey Stolyar, a Research Associate Professor working with my group, and I point out how these units defined upon exchange are actually narrower than “exact sequence variants” (ESVs), which have already been thought of as too narrow by some. We point out that the ecological distinctiveness of lineages in lab-based experimental evolution is often but one or a few mutations, so this seems quite consonant with their findings. It remains unclear whether these populations are equivalent to “species,” but this represents an exciting and refreshingly novel approach to letting genome sequences tell us what we should be paying attention to.
